St. Jude Cloud Ontology
v0.3
Submitted: July 21, 2025
Click here to download.
Version 0.3 of the St. Jude Cloud Disease Ontology (SJC-DO) introduces focused refinements to improve classification accuracy, standardization, and integration of newly available data. These updates build on the structural framework established in v0.2, while incorporating expert feedback, recent publications, and harmonization across PeCan and Genomics Platforms. Major improvements include reclassification of therapy-related AMLs, adoption of standardized gene fusion notation, and structural clean-up of non-leaf nodes and unused classifications across the ontology.
Comprehensive Updates
- Reclassification of Therapy-Related AML Subtypes
- As part of our continued effort to align with expert recommendations and the v0.2 framework, we restructured how therapy-related AML is represented in the ontology to improve clarity and consistency across AML subtypes.
- Standalone therapy-related AML nodes were deprecated to better reflect the evolving classification model, which prioritizes molecular features over clinical context as primary classifiers. Affected samples were reassigned to their corresponding genetic categories, for example, therapy-related AML with NPM1 mutation was reclassified to AML with NPM1 mutation.
- Samples with identifiable but unclassified alterations were reassigned to NEC (Not Elsewhere Classified). For example, SJMDS018205_D1, previously classified as Therapy-related Myelodysplastic Syndrome (TMDS), was reassigned to AMLNEC due to the presence of a rare ZMYM6::STAT3 fusion not represented in existing subtype nodes. Conversely, samples lacking defining molecular features or where no genomic assay was performed - such as SJAML016494_D1 and SJALL016496_D1, both previously classified as Therapy-related AML (TAML) - were reassigned to AMLNOS (Not Otherwise Specified), consistent with v0.2 logic.
- The clinical context of prior treatment exposure was retained using the "post-cytotoxic therapy" qualifier, maintaining traceability of therapy exposure in alignment with WHO guidance.
- Standardization of Gene Fusion and Variant Notation
- To improve consistency and align with community best practices, we adopted the HGNC-recommended double-colon (::) format for all gene fusion names (e.g., ETV6::RUNX1), replacing earlier uses of hyphens or underscores1.
- We ensured all fusions follow 5' to 3' biological orientation to reflect biological directionality. For example, DUX4::IGH (not IGH::DUX4) correctly represents the functional fusion structure, which can impact downstream interpretation and classification.
- Terminology for copy number alterations was standardized using consistent lowercase descriptors such as "amplification", "deletion", "gain", and "loss", eliminating annotation inconsistencies across samples.
- Finally, we limited the use of hyphens to well-characterized structural variants, including KMT2A-PTD, FLT3-ITD, and UBTF-TD, to preserve semantic clarity and reduce mislabeling.
- Ontology Structure and Sample Classification Updates
- As part of routine maintenance and to keep pace with current classification standards, we conducted a focused cleanup of the ontology and processed outstanding sample mappings. We removed redundant and obsolete nodes and reorganized classifications to align with the WHO 2025 hierarchy. For instance, sub-parent terms such as MDMPN were deprecated, while standalone classifications within myeloproliferative neoplasms, such as CML, CNL, JMML, were grouped under the broader Myeloproliferative Neoplasms category (MPN).
- To improve structure consistency, we also added NOS/NEC categories as leaf-level endpoints where none previously existed. For example, cases originally labeled broadly as Rhabdomyosarcoma (RMS) are now routed to RMSNOS if no genomic data is available or RMSNEC if a genetic alteration is present but doesn't match existing subtypes.
Challenges
The main challenge in this update was the ongoing decision-making around how to store and present clinical qualifiers like "post-cytotoxic therapy". While this attribute is retained in the database, we have deferred its inclusion in the public-facing interface pending further discussion on how best to balance clinical value with interface clarity and usability.
Another challenge involved implementing NOS (Not Otherwise Specified) and NEC (Not Elsewhere Classified) categories and properly updating samples that were previously assigned to sub-parent classifications. Samples without genomic subtyping assays were labeled as NOS, while those with rare genetic alterations not fitting established sub-classifications were classified as NEC. For example, samples originally labeled broadly as Rhabdomyosarcoma (RMS) were reclassified as either Rhabdomyosarcoma, NOS if no genetic data was available, or Rhabdomyosarcoma, NEC if genetic alterations were identified but did not align with existing RMS subtypes. This restructuring required detailed review of assay data and careful assignment to preserve classification accuracy.
Future Updates
Looking ahead, we will continue evolving the St. Jude Cloud Disease Ontology (SJC-DO) to better reflect emerging knowledge and ensure consistency across hematologic and solid tumor classifications. Upcoming priorities include expanded work on Acute Lymphoblastic Leukemia/Lymphoma (ALL), with a focus on disentangling classification logic and improving representation of precursor vs. mature lineages. Additional refinements will be made to solid tumor hierarchies as new biomarkers and pediatric-specific subtypes become available.
We also plan to revisit inherited predisposition categories and further standardize how clinical qualifiers, such as treatment history or syndrome associations, are represented within the ontology. Ongoing work will address structural alignment issues, especially around the use of NOS and NEC endpoints, to ensure they are applied uniformly across disease categories.
References
v0.2
Submitted: April 23, 2025
Click here to download.
In our ongoing efforts to update the St. Jude Cloud Disease Ontology (SJC-DO), we have revised the Acute Myeloid Leukemia (AML) classification system to align with the WHO5 framework1, incorporating additional research-relevant classifications informed by a recent study by Umeda et al.2 This review builds on WHO5 to reflect current understanding of AML subtypes, particularly in the context of pediatric AML (pAML). Using this foundation, we re-annotated 1,343 AML samples from the St. Jude Cloud Genomics Platform and PeCan Knowledge Base. These updates were formalized into a hierarchical classification tree, integrated into the broader SJC-DO structure.
Comprehensive Updates
- Refinement of the AML classification system: The AML classification system has undergone significant refinement in v0.2 to better align with the latest standards and provide a structured framework for classification. These updates reduced the ontology to 29 nodes (previously 47). The updated decision tree prioritizes genetic abnormalities, myelodysplasia-related features, and differentiation status, closely mirroring the WHO5 classification.
- Transition from FAB to WHO5: While the FAB classification has been largely deprecated, it remains available as an option when no defining genetic alteration is identified but AML is confirmed. Many diagnoses previously categorized under FAB have been reclassified according to their genetic alterations. For instance, samples initially termed Acute Myelomonocytic Leukemia (AMML) or Acute Megakaryoblastic leukemia (AMKL) that were found to have UBTF tandem duplications are now classified as AML with UBTF Tandem Duplication. Similarly, many FAB-based terms with molecular subtyping such as AMKL and KMT2A rearrangement have been reassigned to AML with KMT2A Rearrangement. When reassignment was not possible due to lack of defining features, they were maintained under the AML Defined by Differentiation category.
- Incorporation of WHO5-aligned structure and inclusion of Myeloid Neoplasms with Germline Predisposition: The classification tree now emphasizes genetic abnormalities, myelodysplasia-related features, and differentiation status. We structured our nodes to mirror the WHO5 hierarchy, including Acute Myeloid Leukemia with Defining Genetic Abnormalities, Acute Myeloid Leukemia, Myelodysplasia-Related, and Acute Myeloid Leukemia Defined by Differentiation. Additionally, a dedicated classification node has been added for Myeloid Leukemia of Down Syndrome under Myeloid Neoplasms with Germline Predisposition, which is distinct from the WHO5 classification.
- Introduction of pediatric-specific categories: Pediatric-enriched drivers like UBTF, GLISr, and GATA1, which are underrepresented or grouped broadly in WHO5, now exist as distinct nodes to reflect their biological relevance in pAML. These alterations are associated with specific subtypes that differ from adult AML in terms of age distribution, prognosis, and treatment response. For example, GLIS family rearrangements are primarily seen in infants with non-Down Syndrome (DS) AMKL and are linked to poor outcomes, while some GATA1 mutations define Myeloid Leukemia of Down Syndrome, a clinically distinct and more favorable subtype3,4. Recognizing these drivers as standalone entities ensures that the classification better reflects the molecular landscape of pAML and supports more accurate research and clinical categorization.
- Use of NOS and NEC categories: We have implemented NOS (Not Otherwise Specified) and NEC (Not Elsewhere Classified) terms to systematically handle assay-dependent limitations or ambiguous cases, preserving structural clarity while expanding classification coverage.
Challenges
The primary challenges in this update stemmed from translating the conceptual framework of WHO5 into a structured ontology. While WHO5 uses a tiered structure—placing some molecularly-defined subtypes at high levels (e.g., AML with NPM1 mutation) and others under umbrella category of AML with other defined genetic alterations—we made a conscious decision to flatten this hierarchy. Rather than grouping molecular subtypes under broad "other" categories, we promoted them to the same structural level of AML with Defining Genetic Abnormalities to improve usability and promote granularity. This flattening required us to rethink how to handle NOS (Not Otherwise Specified) and NEC (Not Elsewhere Classified) designations, particularly in cases where samples lacked definitive markers.
Another key area of complexity involved the transition away from the FAB classification system. We retained the WHO5 strategy by mapping FAB terms without defining genetic alterations under AML Defined by Differentiation, maintaining morphological distinctions where appropriate. FAB-classified cases with known genetic alterations—such as AMKL with KMT2A rearrangement—were reclassified into genetically defined categories, such as AML with KMT2A rearrangement, and grouped under AML with Defining Genetic Abnormalities. In cases where neither genetic nor morphological clarity was available, legacy FAB terms were routed to the NOS bucket. These decisions required careful mapping and manual review, particularly when legacy classifications still held clinical or research relevance but no longer aligned cleanly with modern standards.
Finally, we faced decisions around the appropriate level of granularity for fusion-driven AMLs. While the publication by Umeda et al.2 provides granularity for some rare fusions and not others, it was not intended for standardized sample classification purposes. Given this limitation, we chose to group fusions with well-characterized gene partners—such as NUP98::NSD1 and NUP98::KMT2A—into a single node, AML with NUP98 Rearrangement. Similarly, ETS family fusions such as FUS::FEV, FUS::ERG and related partners, were grouped under a unified node AML with ETS Family Rearrangement. In contrast, rarer and less well-characterized fusions, such as those involving HOX or NPM1 fusion events, were not assigned distinct nodes and were instead routed to the NEC bucket. This strategy balanced clarity and usability while preserving clinically relevant distinctions for pediatric AML.
Future Updates
Building on the advancements in v0.2, we aim to further refine and expand the St. Jude Cloud Disease Ontology to improve scalability and close classification gaps. Our immediate priorities include updating classifications for other hematologic diseases, such as Acute Lymphoid Leukemia/Lymphoma (ALL), incorporating refined classifications for solid tumors, establishing consistent parent-child hierarchies across classifications, and applying the NOS/NEC logic comprehensively across all relevant disease categories. These efforts will ensure the ontology remains aligned with the latest research while maintaining robust pathways for classifying incomplete or ambiguous cases. We also plan to revisit the granularity of fusion-driven AML classifications to establish criteria for separating entities, particularly in cases like NUP98 fusions and other rare rearrangements.
References
- https://tumourclassification.iarc.who.int/
- https://pubmed.ncbi.nlm.nih.gov/38212634/
- https://pubmed.ncbi.nlm.nih.gov/26186939/
- https://pubmed.ncbi.nlm.nih.gov/28112737/
v0.1
Submitted: January 24, 2025
Click here to download.
The St. Jude Cloud Disease Ontology (SJC-DO) is a resource for harmonizing pediatric cancer data. We recently updated the brain tumor classifications to align with the WHO CNS5 Blue Book guidelines 1, 2 and re-annotated 2,587 brain tumor samples in the St. Jude Cloud's Genomics Platform and soon, PeCan Knowledge Base.
Comprehensive Updates
- Reorganized Diffuse High-Grade Glioma (HGG) and Encapsulating Low-Grade Glioma (LGG) categories into a unified parent node: Gliomas, Glioneuronal Tumors, and Neuronal Tumors, including Ependymal Tumor classifications.
- Expanded to include molecular subtypes, enabling more granular classification and precise sample stratification. These are seen in the Pediatric High-Grade Glioma and Pediatric Low-Grade Glioma nodes.
- Re-organized Medulloblastoma into molecularly defined vs histologically defined nodes, given the nature of medulloblastoma's heterogeneity. Also, Medulloblastoma, Group 3 and Medulloblastoma, Group 4 have been re-classified under Medulloblastoma, non-WNT/non-SHH to align with WHO CNS5 guidelines.
- We have new defined classifications, such as CIC-rearranged Sarcoma and Dysembryoplastic Neuroepithelial Tumors.
- Added entries for NOS/NEC, addressing assay-dependent classification gaps and ensuring that samples are systematically categorized even when data is incomplete.
- The v0.1 ontology now consists of 80 nodes (reduced from 94) and expands to five layers (see Figure 1 below).
Challenges
The hierarchical presentation of diseases in the CNS5 guidelines lent itself readily to translation into our tree structure. However, there were cases where the modeling was not completely straightforward. Here we describe some of the challenges and the approach we took to each.
The WHO CNS5 blue book lays out definitions for not-otherwise-specified (NOS) and not-elsewhere-classified (NEC) that clearly distinguish the terms and allow application of them to any disease term having subtypes. However, there is an explicit combined NOS/NEC term for embryonal tumors, CNS Embryonal Tumor NEC/NOS. To ensure consistent handling of NOS/NEC terms in our ontology, we added explicit and separate NOS and NEC nodes for every non-leaf node. In the case of CNS Embryonal Tumor NEC/NOS, we included this node but treated it as an NEC node when biomarker data was available. In cases where biomarker information was unavailable and a sample could not be mapped to Embryonal Tumor subtypes, those samples were mapped to Embryonal Tumors, NOS, adhering to the comprehensive NOS/NEC framework.
The tree structure itself presents challenges, such as the non-nested presentation for Posterior Fossa and subtypes, Posterior Fossa A and Posterior Fossa B. We elected to model these as three sibling nodes in our tree, mirroring the presentation in the blue book. Similarly, Supratentorial Ependymoma and its non-nested ZFTA and YAP1 subtypes were modeled as siblings. While this structure aligns with the presentation of these terms in WHO CNS5, in a future version we will likely nest these subtypes under the parent nodes. When classifying our posterior fossa samples, because methylation profiling data is not yet available to differentiate between A and B, we mapped these samples directly to Posterior Fossa. This approach is informative, as it distinguishes them from other ependymal tumor types like Myxopapillary or Supratentorial. Until methylation profiling data becomes available, all Posterior Fossa samples remain classified under the broader Posterior Fossa category.
In other cases, such as Atypical Teratoid/Rhabdoid Tumor (ATRT), the blue book entry describes subtypes but embeds the subtype classifications within the descriptive text of the entry, reducing clarity. Following this structure, we mapped ATRT subtypes similarly without creating separate child nodes. Given the small sample size for these subtypes, we opted to roll them back to the broader ATRT term. Subtype biomarkers are provided if further classification becomes necessary in the future.
Embryonal tumors, particularly Medulloblastoma, introduce complexity with separate molecular and histological branches, raising uncertainty about whether these branches inherently indicate NOS/NEC logic. To address this, we mapped samples to child leaf nodes, treating parent nodes as aggregates. Samples were classified into one of the four molecularly defined nodes based on subtype biomarker data or directly to the "Medulloblastoma, Histologically Defined" node, such as samples previously classified as "Large Cell Medulloblastoma" in our v0 ontology.
In conclusion, while we aimed to align closely with the WHO CNS5 structure to maintain consistency, there were cases that required making modeling decisions that involved trade-offs between competing priorities. Time may reveal better approaches to some of these challenges, yielding updates to the ontology. Additionally, as new cases emerge and the transition to CNS6 occurs, the ontology will need to evolve further.
Future Updates
Building on the advancements in v0.1, we aim to further refine and expand the St. Jude Cloud Disease Ontology to address gaps and enhance scalability. Immediate priorities include updating hematological diseases, particularly Acute Myeloid Leukemia (AML), and incorporating updated solid tumor classifications. These efforts will focus on creating consistent parent-child hierarchies and applying the NOS/NEC logic comprehensively across these domains.
In tandem, we are developing a pilot for an Encyclopedia of Composable Characteristics (ECC). This publicly available resource found on GitHub 3, 4 will prioritize the decoupling of key molecular, histological, and other evidence into independently assignable attributes. Future versions of the ontology will include rigorous mappings from ECC to ontology terms. By adopting this composable approach, we aim to minimize disruptions during future updates, support dynamic classifications, and empower other ontologies to be applied.
With each iteration, we aim to align the ontology more closely with cutting-edge research needs while maintaining robust pathways for incomplete or ambiguous cases. This commitment to continuous improvement ensures the St. Jude Cloud Disease Ontology (SJC-DO) remains a vital resource for the global research community, enabling seamless data harmonization and driving new discoveries in pediatric oncology.
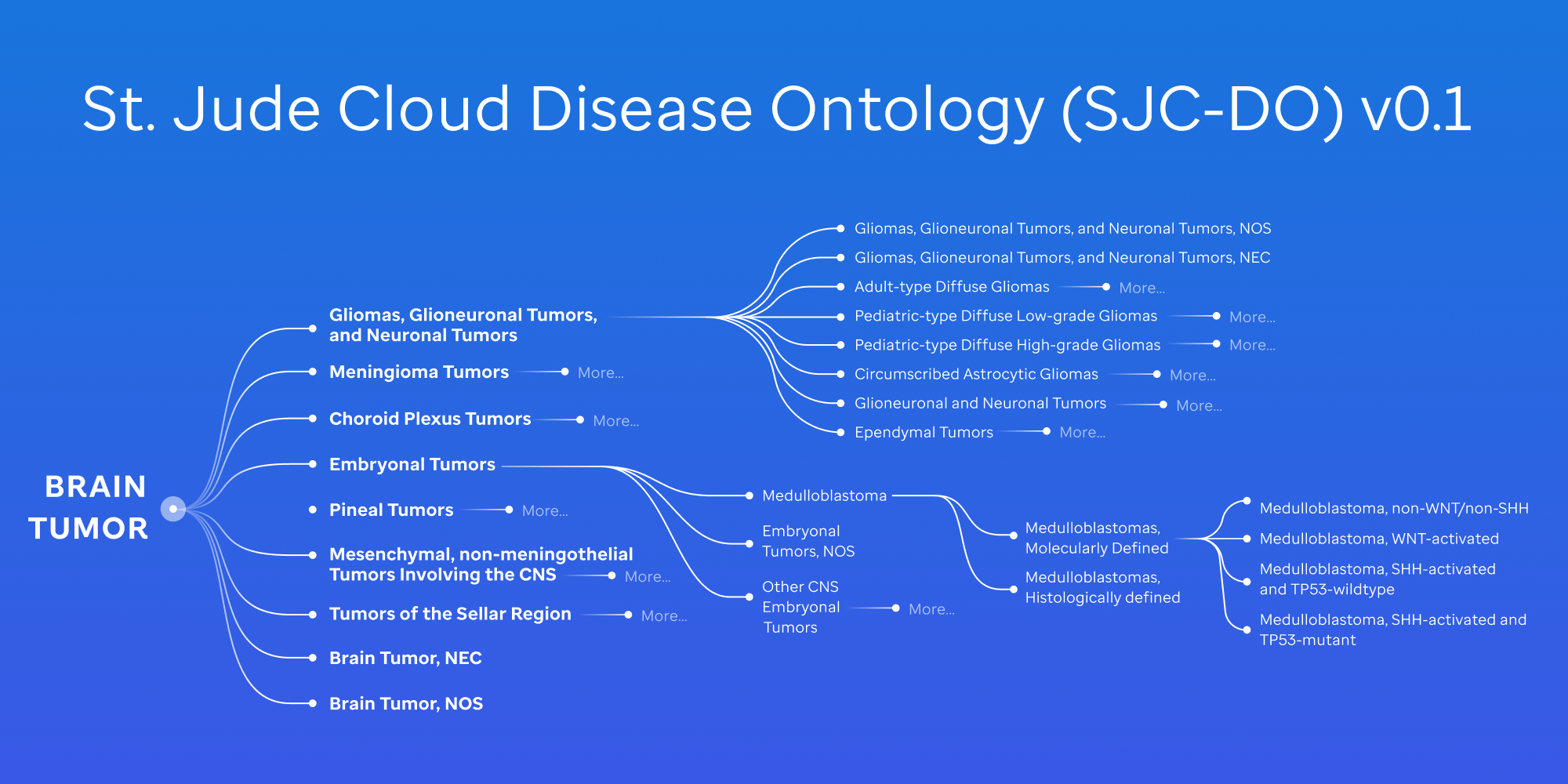
Figure 1: St. Jude Cloud Disease Ontology (SJC-DO) v0.1 aligning to the WHO CNS5 guidelines.
References
- https://tumourclassification.iarc.who.int/
- https://pmc.ncbi.nlm.nih.gov/articles/PMC8328013/
- https://github.com/stjudecloud/ecc
- https://github.com/stjudecloud/ontology
v0.0
Submitted: May 14, 2024
Click here to download.
Ontologies designed for disease classification have redefined our understanding of diseases by providing a hierarchical structure of complex biomedical data. In cancer research, they are critical for data sharing, integration, and collaboration among researchers. However, existing ontologies on pediatric cancer classification are limited. The World Health Organization (WHO) and OncoTree primarily focus on adult cancers while leaving gaps in many pediatric cancer subtypes driven by molecular etiology presented in recent scientific literature. To enable data sharing and integration of the whole-genome, whole-exome and RNA-seq data generated from 13,956 cases of pediatric cancer and long-term survivors on St. Jude Cloud, we recognized the significance of such gaps and initiated the development of a tailored disease ontology to address this issue.
Principles
Our goal is to develop a pediatric-centric framework with the capability of integrating new research findings, including those involving rare molecular drivers. Currently, we focus exclusively on pediatric cancer but will consider extension to other childhood catastrophic diseases, such as Bone Marrow Failure and Sickle Cell disease. Our framework is designed to integrate molecular, pathological, and histological features by leveraging existing efforts from OncoTree, WHO, and community knowledge.
Methods/Details
To achieve these principles, we evaluated existing ontologies, including:
- OncoTree
- International Classification of Diseases for Oncology (ICD-O)
- The World Health Organization (WHO) Hematological and CNS classifications
Our primary design is based on OncoTree due to its cancer-focused approach, structure for starting at tissue and breaking out into diseases, and alignment with our guiding principles. However, there have been deviations which initiated the tailored ontology that applies across St. Jude Cloud (Figure 1).
Key Structural Changes
- Adjustment in Hematological Diseases: Distinctions between leukemia and lymphoma were introduced, addressing a gap in OncoTree's applicability to the pediatric domain.
- Exclusion of Adult-Specific Terms: We omitted terms exclusive to adult diseases, such as breast cancer and lung cancer (e.g. small cell lung cancer (SCLC)) ensuring our ontology's focus primarily remains tailored to pediatric oncology.
- Expansion for Recently Discovered Molecular Drivers: Recognizing the prominence of new molecular drivers discovered by genome-wide profiling of pediatric diseases, we expanded our ontology to include additional nodes to reflect the current knowledge. For instance:
- B-Cell Acute Lymphoblastic Leukemia (BALL)1 was subdivided into 28 distinct subtypes, a considerable increase from OncoTree's original nine.
- Incorporating newly discovered molecular drivers such as DUX42, MEF2D3 NUMT1 or BCL11B4 rearrangements.
- T-Cell Acute Lymphoblastic Leukemia (TALL)5 was classified into 10 distinct subtypes, up from two in OncoTree.
- Including commonly activated transcriptional regulators including those oncogenes defining T-ALL subgroups - TAL1, TLX1, TLX3, and NKX2-1.5
- B-Cell Acute Lymphoblastic Leukemia (BALL)1 was subdivided into 28 distinct subtypes, a considerable increase from OncoTree's original nine.
Current Status
To date, the development was primarily motivated by omics data that was being uploaded to the St. Jude Cloud platform every month for community data sharing. At this cadence, the ontology framework represents an evolving architecture as it is continuously being refined as new data is curated. In addition to the incremental updates, major revision has been planned, with current examples described below. This is to align with recent publications, to work closely with institutional experts who are heavily involved with developing the WHO classifications, thereby updating CNS tumor classification updates from WHO CNS5, to make recent updates from OncoTree.
Current Focus
Review of Glioma Tumors6:
- There's a notable shift in classifying diffuse intrinsic pontine glioma to midline glioma, reflecting evolving understanding and diagnostic criteria noted by the WHO CNS5 guidelines.
- Additionally, our disease ontology's inclusion of modifiers such as anaplastic or diffuse diverges from the recent WHO CNS5 classification updates for grading, particularly concerning tumors like astrocytoma and glioblastoma.
Review of Embryonal Tumors6:
- Recent studies advocate for revisiting the classification of embryonal tumors, for example medulloblastoma groups 3 and 4. Proposals suggest annotating them as Medulloblastoma, non-WNT/non-SHH7,8 emphasizing molecular distinctions over histological classifications.
- Given the heterogeneity nature of medulloblastoma, there is a new term, histologically defined, that should be evaluated and employed for subtypes such as large cell/anaplastic or desmoplastic/nodular medulloblastoma.
Review of Solid Tumors9:
- Explore merging subtypes such as osteoblastic osteosarcoma and chondroblastic osteosarcoma under the umbrella of osteosarcoma, aligning with evolving research insights.
Conclusion
Our disease ontology is integral to various applications within St. Jude Cloud, driving initiatives like the Genomics Platform and Pediatric Knowledge Base (PeCan). However, its growth and effectiveness rely on community involvement. The current ontology framework has been developed with the input from pathologists and researchers involved in molecular subtyping. We welcome additional input and collaboration from researchers and clinicians to ensure its ongoing improvement and relevance to pediatric oncology, ultimately contributing to better outcomes for children facing cancer and catastrophic diseases.
Contact: For inquiries, collaborative opportunities, or to provide feedback on improving the St. Jude Cloud disease ontology, please contact support@stjude.cloud.
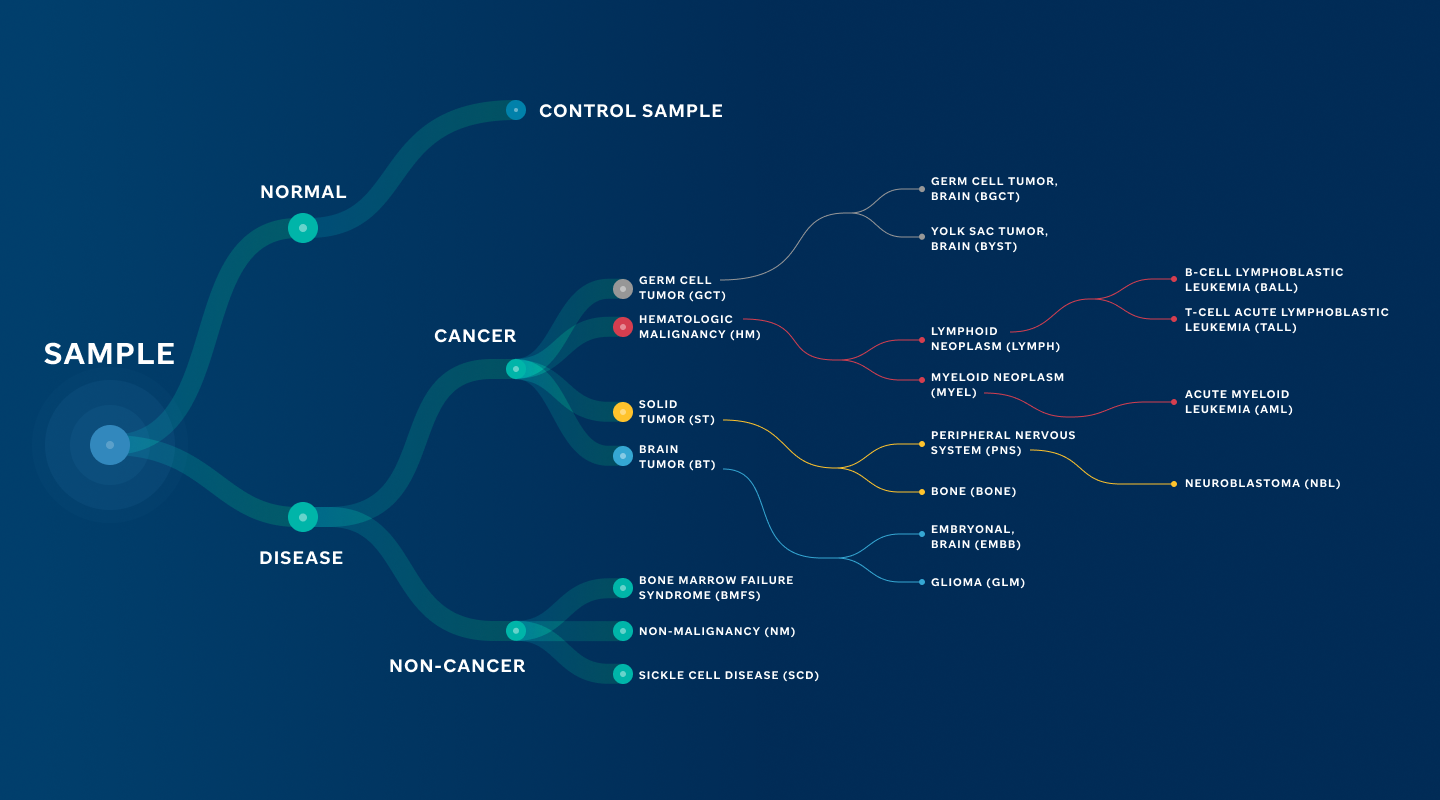
Figure 1: St. Jude Cloud Disease Ontology. High-level overview of the ontology that supports applications in St. Jude Cloud.
References
- https://pubmed.ncbi.nlm.nih.gov/36050548/
- https://pubmed.ncbi.nlm.nih.gov/27776115/
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5105166/
- https://pubmed.ncbi.nlm.nih.gov/36050548/
- https://pubmed.ncbi.nlm.nih.gov/28671688/
- https://academic.oup.com/neuro-oncology/article/23/8/1231/6311214
- https://www.biorxiv.org/content/10.1101/2024.02.09.579680v1.full
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8833659/
- https://ascopubs.org/doi/full/10.1200/CCI.20.00108
Metadata and Clinical Information
Working With Our Data
In this overview, we will explain how to manage your data request(s) from St. Jude Cloud's genomics platform My Dashboard page and how to access and manage your data (once it has been vended to you) from within a DNAnexus project. The DNAnexus genomic ecosystem is the backbone for the computation and storage in St. Jude Cloud. This means that each data request in St. Jude Cloud corresponds to a project in DNAnexus. If you'd like, you can read an introduction to the DNAnexus ecosystem here. If you haven't already, follow this guide to request access to St. Jude data in this secure cloud ecosystem.